From Past to Present
Sequencing technologies have revolutionized the field of genomics, enabling genetic information to be decoded with unprecedented speed and accuracy. These advanced techniques allow the precise order of nucleotides in a cell’s DNA or RNA to be determined, providing crucial insights into biological processes, disease mechanisms, evolutionary patterns, and more. Over the past decades, multiple sequencing technologies have emerged, each bringing its own strengths and limitations.
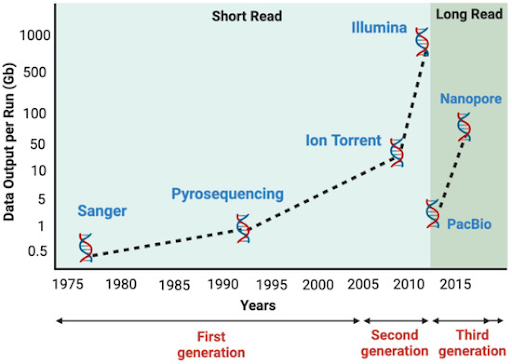
First generation: Sanger sequencing
Two pioneering methods of sequencing were developed in the late 1970s, the Maxam-Gilbert and Sanger methods. The Maxam-Gilbert method, which is based on chemical degradation, is not widely used due to various disadvantages such as high toxicity and errors during chemical cleavage. Sanger sequencing, developed by Frederick Sanger based on the chain elongation termination principle, was the first widely adopted sequencing method. It relies on the random incorporation of chain-terminating dideoxynucleotides (ddNTPs) by DNA polymerase during replication, which is then visualized with gel electrophoresis. This method was subsequently improved with the introduction of fluorescently labelled ddNTPs and capillary electrophoresis. Sanger sequencing remains the gold standard today and is highly accurate for up to about 1,000 base pairs, particularly in complex regions involving repeats and secondary structures. While it is still widely used today for small scale sequencing projects and relied upon as an orthogonal method to verify variants detected by other sequencing technologies, its labor-intensive process, high cost, long turnaround time, and low throughput limit its use in large scale genomic studies or routine diagnostics. Additionally, Sanger sequencing can only detect a minimum allele frequency of 15 to 20%, limiting its ability in detecting rare variants.
Second generation: Next-generation sequencing
The introduction of next-generation sequencing (NGS) in the early 2000s marked a transformative leap in sequencing technology. Unlike Sanger sequencing, NGS uses a massively parallel approach to sequence millions of DNA fragments simultaneously. Because of its high throughput capability, NGS dramatically reduces the time and cost when sequencing large areas of the genome or analyzing a broad panel of genes. Moreover, NGS provides higher depth of coverage, which increases the accuracy of detecting low-frequency variants in heterogenous samples and requires only a small amount of sample, making NGS highly suited for routine diagnostics. Since NGS typically generates shorter read lengths (up to about 600 base pairs), which are then computationally assembled into longer sequences, this process can make it challenging to resolve repetitive or complex genomics regions accurately.
Third generation: Long-read sequencing
To address the limitations of short-read sequencing, long-read sequencing that can produce longer read lengths (up to about 30,000 base pairs) was developed. Because longer reads can cover larger portions of the genome in a single sequence, they excel at characterizing repetitive and complex genomics regions, resolving large structural variations, phasing haplotypes, and assembling genomes de novo. While long-read sequencing is not yet routinely used in cancer diagnostics, it offers significant advantages in other areas such as studying rare and inherited diseases, many of which are caused by complex or large genetic variations that might be difficult to detect with short-read technologies. Although this sequencing method is associated with higher error rates and costs compared to NGS, technological advancements in chemistries and bioinformatics tools are closing the gap. Sequencing technologies have transformed our ability to explore the genetic code, from the early days of Sanger sequencing to the present era of long-read sequencing. Each technology offers distinct strengths: Sanger sequencing offers precision for small targets, NGS delivers massive scale and efficiency, and long-read sequencing provides high resolution of complex genomic features. Looking ahead, the future of sequencing lies in improving the performance of these technologies and making them more accessible. As innovation continues, these tools will play an increasingly pivotal role in enabling new scientific discovery and driving the next generation of healthcare.
Canary Oncoceutics has a steadfast commitment to three fundamental pillars: advancing scientific knowledge, fostering collaboration, and ultimately, enhancing the lives of cancer patients worldwide. From cutting-edge research to impactful clinical advancements, Canary Oncoceutics aims to illuminate the transformative potential of tailored cancer treatments. Join us on this journey towards a future where every cancer patient receives personalized, effective treatment tailored to their unique needs.