Introduction
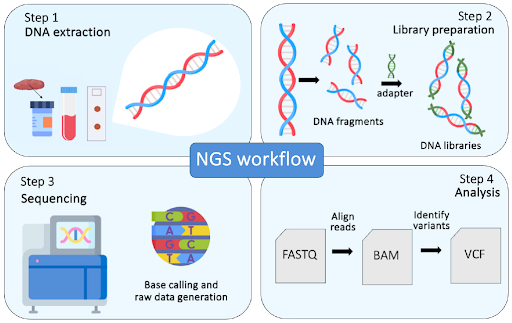
Next-generation sequencing (NGS) is a revolutionary tool in cancer research and diagnostics that enables comprehensive analysis of genetic alterations with an unprecedented speed, accuracy, and scalability, in a single test. By identifying various mutations such as insertions, deletions, copy number variations and gene fusions across multiple cancer-associated genes, this information helps in predicting cancer risk, diagnosing cancer, selecting appropriate therapies, monitoring treatment response and disease progression. The NGS workflow is a complex process that typically involves four key steps: nucleic acid isolation, library preparation, sequencing and data analysis.
Nucleic acid isolation
The first step in the NGS workflow is the collection of biological samples, which could be blood, saliva, tissue, or other biological material. Once the sample is obtained, nucleic acids (DNA or RNA) are isolated and purified using specialized commercial kits. Since the yield, purity and quality of the isolated nucleic acids are crucial for the success of downstream processes, these parameters must be analyzed and optimized as part of the quality control process before proceeding to library preparation.
Library preparation
In this step, the isolated and purified nucleic acids undergo processing so that they can be recognized by the sequencing platform. Although library preparation protocols vary depending on the methods and technologies used, the nucleic acids are often first fragmented into smaller pieces to enable massively parallel sequencing. The ends of these fragments are then ligated with specific adapters that allow for the fragments to be recognized and amplified during sequencing. For RNA sequencing, the RNA is first reverse-transcribed into cDNA, which is then fragmented and prepared in a similar manner. If needed, unique barcodes (short, known sequences that are added to the adapters), can be used to tag each sample. This allows multiple samples to be pooled together in a single sequencing run while still enabling them to be distinguished after sequencing (this is known as multiplexing). Following adapter ligation, the individual libraries are quantified, normalized and pooled together. This quality control step helps to ensure consistent data output, high sequencing quality, as well as efficient use of the sequencing reagents and flow cells.
Sequencing
Once the pooled library is ready, it is loaded onto the sequencer. Depending on the sequencing platform, sequencing can occur through various technologies, such as sequencing-by-synthesis (Illumina), semiconductor sequencing (Thermo Fisher Scientific) or DNA nanoball sequencing (MGI Tech). During sequencing, the DNA or cDNA fragments are read base by base by the sequencer (this process is known as base calling), which generates raw data in the form of short sequences called reads. These reads are generated rapidly and in parallel, with up to billions of fragments sequenced in a single run.
Data analysis
After sequencing is completed, the raw data containing the nucleotide sequences (reads), along with quality scores (indicating the confidence of the base call), and other relevant information is generated. The raw data then undergoes processing and quality control such as adapter trimming, demultiplexing (separating reads from different samples based on sample-specific barcodes), and removal of low-quality bases. Next, the high-quality, sample-specific reads are aligned to a reference genome using alignment tools. After alignment, the data can then be analyzed. For DNA sequencing, this may involve variant calling (identifying mutations), while for RNA sequencing, this may include transcript quantification or differential expression analysis. The final step involves interpreting the results using biological databases, established guidelines, and annotation tools to assess the functional or clinical significance of the findings. The findings are typically presented in a comprehensive report that provides various insights, such as genetic predispositions, disease mechanisms, potential therapeutic targets, or therapy options. In summary, the NGS workflow comprises multiple complex steps, each of which is accompanied by rigorous quality control measures to ensure that the resulting data is both accurate and of high-quality. Undoubtedly, NGS has now become a pivotal tool in a wide range of applications, from basic research to clinical diagnostics. In precision oncology, NGS plays a crucial role by enabling personalized treatment and monitoring through comprehensive genomic profiling. This approach aligns with the ultimate goal of precision oncology, where care is tailored to meet the unique needs of each individual.
Canary Oncoceutics has a steadfast commitment to three fundamental pillars: advancing scientific knowledge, fostering collaboration, and ultimately, enhancing the lives of cancer patients worldwide. From cutting-edge research to impactful clinical advancements, Canary Oncoceutics aims to illuminate the transformative potential of tailored cancer treatments. Join us on this journey towards a future where every cancer patient receives personalized, effective treatment tailored to their unique needs.